Haemophilus influenzae is the gram negative cocco bacilli which was first isolated by Richard Pfeiffer during the influenza pandemic of 1890. It was previously called Pfeiffer’s bacillus or Bacillus influenzae. It was mistakenly thought to be the cause of the disease influenza, and it was named accordingly. More than 19 Haemophilus species can be found till now. Some of them are Haemophilus Haemolyticus, Haemophilus parainfluenzae, Haemophilus ducreyi, Haemophilus aprophilus, Haemophilus influenzae and so on. H. influenzae is the major pathogen of the genus Haemophilus. It may be either capsulated (typeable) or non-capsulated (non-typeable). Based on capsular polysaccharide, it is of six serotypes, similarly, based on three biochemical tests i.e. indole production, urease activity and ornithine decarboxylase, H. influenzae is of eight biotypes. Majority of clinical isolates are biotypes I, II and III, where invasive Hib strains are of biotype I.
Classification
Table 1: Classification of Haemophilus influenzae
| Kingdom | Bacteria |
| Phylum | Proteobacteria |
| Class | Gamma Proteobacteria |
| Family | Pasteurellaceae |
| Genus | Haemophilus |
| Species | influenza |
Morphology and Microscopy
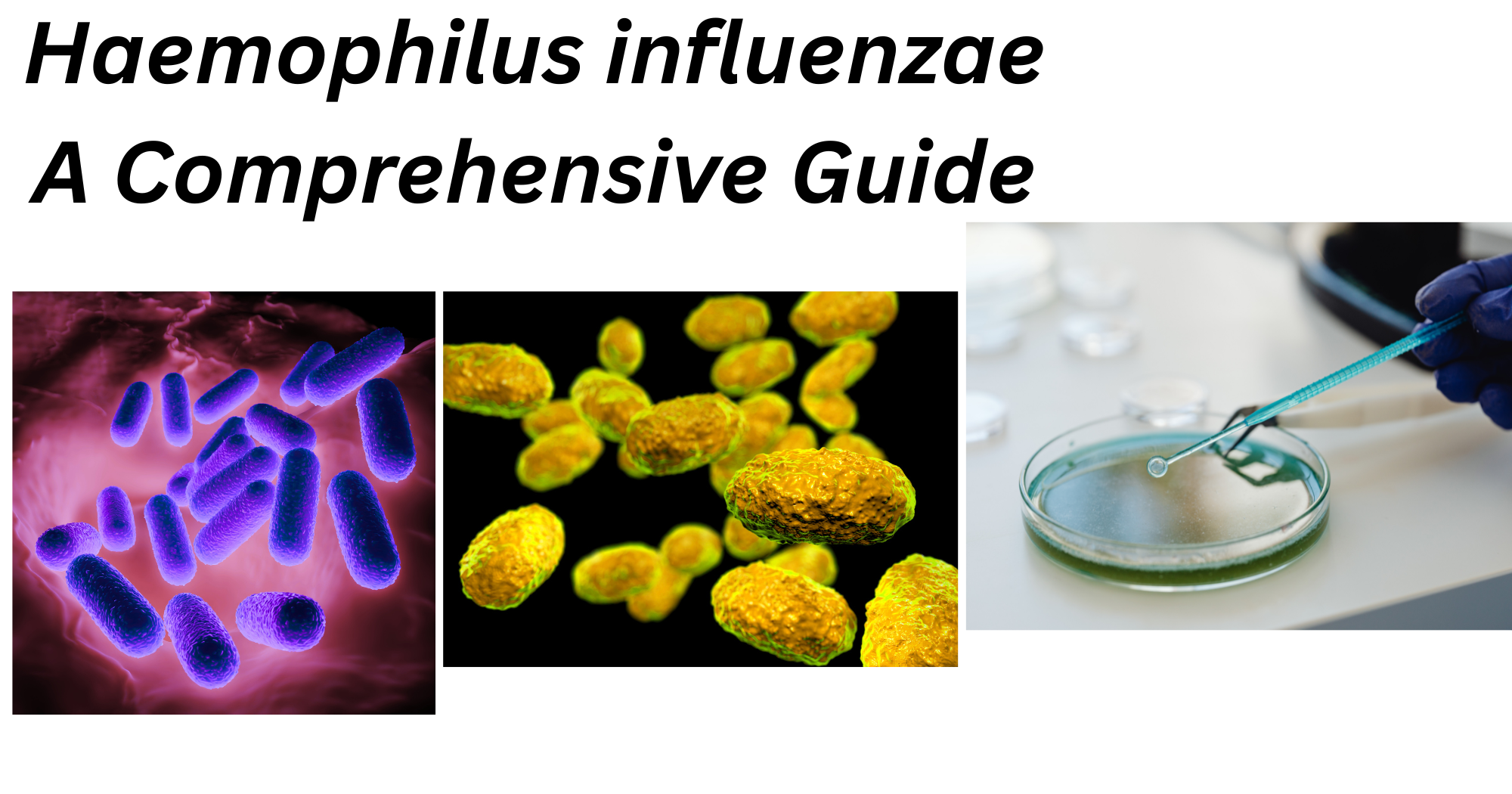
- Gram-negative pleomorphic or coccobacilli
- Small in size, usually 0.3 μm to 1 μm
- Long , thread and occasionally filamentous
- Non motile
- Non sporing
- Aerobic and Facultative anaerobe
- Capsulated (capsules play major role in pathogenesis)
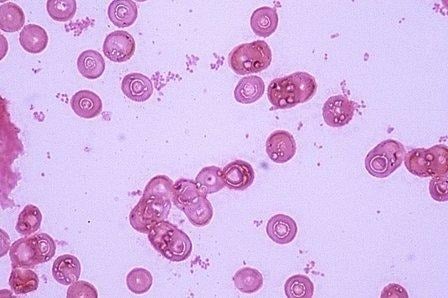
Fig 1: Microscopic structure of H. influenzae
Image source:
https://microbiologylearning.weebly.com/gram-negative-coccobacilli.html
Cultural and Growth Characteristics
- H. influenzae grows well in chocolate agar where colonies are moist, smooth and gray.
- It produces colonies of 0.5mm in diameter on horse Blood Agar when incubated at 37°C for 24 hrs.
- Also, when sub-cultured on a transparent medium such as Levinthal agar then colonies of capsulated strains show iridescence. It is seen as different shades of red, orange, green and blue that change with angle of observation.
- Opaque, mucoid and smooth kinds of colonies are formed by the capsulated strains and the diameter observed is 3-4mm.
- Fastidious and facultative anaerobes can also grow in 5% CO2.
- The optimum temperature needed for H. influenzae to grow is 35-37°C and optimum pH is 7.6.
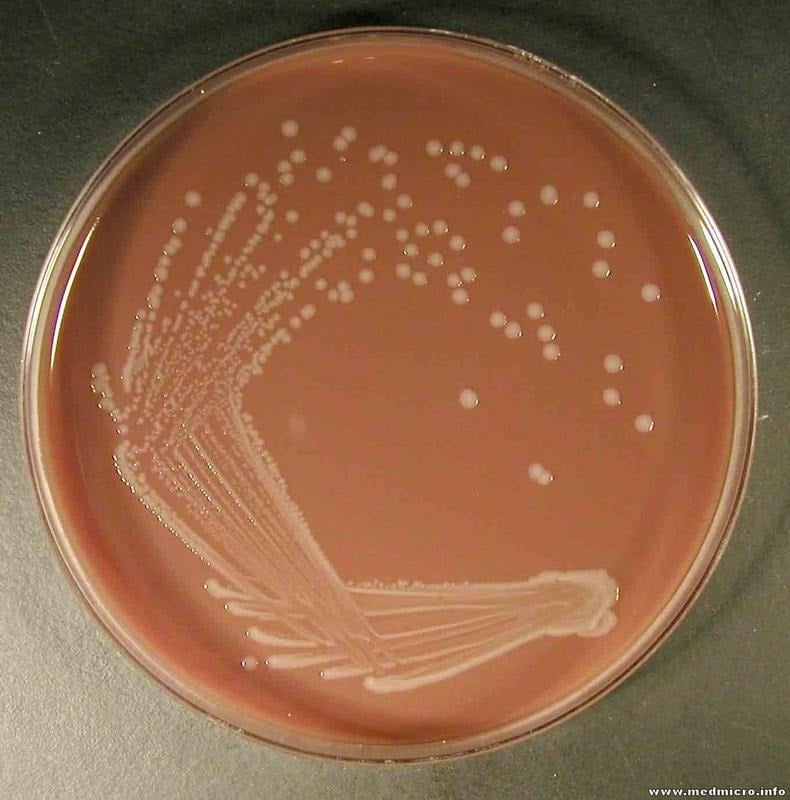
Fig 2: H. influenzae in Chocolate agar
Image source:
https://microbiologylearning.weebly.com/gram-negative-coccobacilli.html
Biochemical and Identification Tests
Table 2: Biochemical tests for identification
| Test | Results |
| Catalase | Positive |
| Oxidase | Positive |
| Indole | Varies |
| Urease | Varies |
| Glucose | Positive |
| Fructose | Negative |
| Xylose | Positive |
| Lactose | Negative |
Virulence Factors and Pathogenesis
Some of the virulence factors and their roles in pathogenesis are described below.
- Capsule:
In Hib, capsular polysaccharide is composed of phosphoribosoryl ribitol phosphate (PRP) which has alphagocytic properties and is highly associated with virulence.
- Membrane Lipooligosachharide:
It is responsible for bacterial attachment, invasiveness and paralysis of ciliated respiratory epithelium.
- There are involvement of other cell envelope factors too in facilitating the attachment to host cells.
- Also, secretory antibodies are probably inactivated by IgA protease.
- Pili and other cell factors help in attachment to host cells.
Pathogenesis of H. influenzae includes two phases. They are:
- Portal of entry: Upper respiratory tract, especially the nasopharynx is the entry route of the H. influenza (usually all strains).
- Dissemination: When H. influenzae enters the body, its type b penetrates the nasopharyngeal epithelium. It either spreads through the blood stream or directly to the meninges. It can transmit from sinuses to meninges and is only second to Streptococcus pneumoniae as the most common cause of recurrent meningitis. People who suffer from chronic otitis media, head injury or CSF leak are highly at risk of getting infected by H. influenzae.
Epidemiology and Transmission
Usually, reservoirs of H. influenzae are human. There is no animal reservoir so we can say that H. influenzae are solely human pathogens. Non-capsulated strains of H. influenzae carried by healthy children are around 60-80%, healthy adults carry around 35%. Asymptomatic carriers of capsulated H. influenzae type b by healthy children are around 2-3%. It can transmit through person to person spread by contaminated respiratory droplets. Also, certain infections can be caused by a person’s endogenous strains.
Clinical Manifestations
The diseases that are caused by H. influenzae can be divided into two groups i.e. invasive and noninvasive. The common invasive and non-invasive groups are described below:
- Invasive infection: It includes diseases like meningitis, acute epiglottitis, bacteriaemia, suppurative lesions, pneumonia etc. Few of them are described below:
- Meningitis: most common in children between 6 months and 2 years of age.
- Acute epiglottitis: It is an acute inflammation of the epiglottis. The swelling of cellulitis and supraglottic tissues are seen which makes it one of the life threatening diseases that are found in the small children (age 2-3 years).
- Bacteraemia: Children who become febrile can be infected by the H. influenzae type b which causes bacteraemia. When these bacteria transmit from blood then it may lead to suppurative lesions. Sometimes, neonates can also be infected by non-capsulated H. influenzae causing septicaemia and meningitis.
- Pneumonia: Lobar pneumonia can be caused by this bacteria, mainly in older patients, chronic alcoholics and smokers etc.
- Non-invasive infections: Many local infections such as otitis media, sinusitis, conjunctivitis etc can be caused by both capsulated or non capsulated strains of H. influenzae.
Laboratory Diagnosis:
The laboratory diagnosis of H. influenzae are listed below:
- Specimens: specimens depend on the site of infection and include CSF, Nasopharyngeal swab, Blood, Pus, Sputum, Urine (rarely in case of UTI), Joint aspirates.
Note: All the specimens should be kept at room temperature and processed within 10 minutes of collection.
- Direct microscopy: Gram stain: Gram-negative coccobacilli
- Gram stained smear of sediment obtained after centrifugation of CSF at 2000rpm for 10 min reveals that H. influenzae.
- Capsules of capsulated strains are not easily shown on these tiny organisms except serologically by quelling reaction.
- Culture:
- Specimens should be cultured as soon as possible.
- It grows better aerobically (CO2 enriched) than anaerobically.
- Culture media that can be used for H. influenzae are Chocolate agar, Levinthal agar, Levinthal broth, BHI agar supplemented with X and V factor and BHI broth supplemented with X and V factor.
- After the inoculation of the specimen into CA, the plate should be incubated in a 5-10% CO2 enriched atmosphere (commonly used is Candle jar) at 37°C for 24hrs.
- It gives round, convex colonies with an opaque zone near the centre.
- Satellitism phenomena:
- H. influenzae requires X and V factors for their growth.
- X factor is for the synthesis of respiratory enzymes like: cytochrome, catalase and peroxidase, a protoporphyrin IX (haematin, a precursor of haemin) is required.
- V factor- NAD is used as an electron carrier in the organism’s oxidation reduction system.
- Blood agar that is regularly used contains X factor but lacks V factor due to presence of real intact cells in the medium.
- When blood agar is heated to 75°C more V facters are released, so chocolate agar is best for the growth of this organism.
- H. influenzae produces colonies on horse BA of about o.5mm in diameter. However, if S. aureus which produces V factor in excess of its own need is cultured on BA plate with H. influenzae, other substances and V factor produced by S. auerus help the growth of H. influenzae which forms the basis of satellitism test which is the simplest way of recognizing H. influenzae regularly.
- S. pneumoniae also produce V factor and cause H. influenzae to show satellism.
- A loopful of suspected Haemophilus colonies is mixed in about 2ml of sterile peptone water or normal saline.
- Then by using a sterile swab, inoculate the organism suspension on a plate of NA and BA.
- A pure culture of S. aureus is streaked across each inoculated plate.
- Both plates are incubated in CO2 enriched atmosphere at 35-37°C overnight.
- If there is growth on BA plate but not on NA plate and colonies near columns of S. aureus growth are than those farthest from it then it is assumed to be H.influenzae.
Fig 3: Haemophilus influenzae and Staphylococcus aureus.
Source:
https://microbiologylearning.weebly.com/gram-negative-coccobacilli.html
Table 3: Satellitism Phenomenon
| Haemophilus | X factor | V factor | Xylose |
| H. influenzae | positive | positive | positive |
| H. aegyptius | positive | positive | negative |
| H. parainfluenzae | negative | positive | negative |
Quelling technique
- The test is performed by placing a drop of CSF, broth or saline suspension of the test organism on each end of a slide.
- A loopful of H. influenzae type b antiserum is mixed with one drop of saline.
- Both preparations are then covered with a coverslip and looked under oil immersion.
- The organism can be identified as Hib if rods or coccobacilli surrounded by clearly defined capsules are seen in the prepared antiserum when compared with saline preparation.
- Because the antibody is deposited on the bacterial cell wall the capsules are seen in the antiserum preparation.
- If each oil immersion field holds 10-50 orgs then best reactions are obtained.
Antigen detection
Type b polysaccharide ag (PRP capsules) of H. influenzae in body fluids (eg: CSF or pus) can be done using latex agglutination and coagglutination. Less than 1ng/ml of PRP can be detected in latex agglutination tests. Some of the molecular techniques such as Polymerase chain reaction are mostly used nowadays for the detection of H. influenzae.
Treatments
Antibiotics such as Ampicillin, Chloramphenicol, Cefotaxime, Ceftazidime etc are effective against H. infuenzae. However, 25-30% invasive strains showed resistance to most of the antibiotics used so Antibiotic susceptibility testing must be done before prescription. Cephalosporins and Chloramphenicol (third generation) can be used in case penicillin resistance is observed.
Prevention and Control
- PRP-conjugate vaccines (capsular antigen) can be used for long term protection.
- The major control measures should be used by healthcare professionals by strictly following hospital safety protocols.
- Contact between infected patients and other individuals should be maintained.
Conclusion
In conclusion, H. influenzae holds the major clinical impact because of its virulence, persistence, and antibiotic resistance, mainly in vulnerable patients’ group therefore, it is an important gram-negative coccobacillus that needs our attention towards.
References
Khattak ZE, Anjum F. Haemophilus influenzae Infection. [Updated 2023 Apr 27]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK562176/
Mardh P A, Taylor-Robinson D eds 1984 Bacterial Vaginosis. Almqvist and Wiksell.
Jordens J Z, Slack MPE 1995 Haemophilus influenzae then and now. Eur J Clin Microbiol 14:935-948.
