Among 191 species of genus Pseudomonas, only more than 25 have been identified in association with humans, with Pseudomonas aeruginosa one of them. P. aeruginosa is an opportunistic pathogen commonly found in the environment mainly in soil and water, but it is also regularly found on plants and sometimes on animals, including humans. This bacterium is known for causing a wide range of infections, from localized infections after surgery or burns to fatal bacteremia. It also causes catheter-associated urinary tract infections, necrotizing pneumonia, severe eye and middle-ear infections, mainly in children and rarely causes meningitis after lumbar puncture, endocarditis after cardiac surgery. It has been linked with some diarrheal disease cases as well. The first case of P. aeruginosa was reported in 1890, after that it has become an important cause of Gram-negative bacteremia, resulting in 15% of cases with an overall mortality of around 50%.

Taxonomy and Classification
Table 1: Classification of P. aeruginosa
| Kingdom | Bacteria |
| Phylum | Proteobacteria |
| Class | Gammaproteobacteria |
| Order | Pseudomonadales |
| Family | Pseudomonadaceae |
| Genus | Pseudomonas |
| Species | aeruginosa |
- Based on the rRNA/DNA hybridization studies, Pseudomonas can be divided into rRNA homology groups, each of which is represented by at least a separate genus.
Structure and Morphology
- Average size: 0.5 – 0.8 µm X 1.5 – 3.0 µm
- Gram-negatives often occur as single cells or in paired rods.
- Obligate aerobe
- Encapsulated
- Non-spore forming
- Do not ferment carbohydrates
- Colour: Pink/ Red
- P. aeruginosa are motile with a single flagellum inserted at the tip of the cell, either polar or subpolar.
- The flagella yield heat-labile antigens (H antigens) acting as a virulence factor in pathogenic strains.
- Polar fimbriae or pili are also found in some strains of P. aeruginosa.
- These pili are usually 6 nm wide and act as a receptor for various phages and are even retractile.
- The cell envelope has three distinct layers; outer membrane, inner cytoplasmic membrane, and the peptidoglycan layer.
- The cell membrane of the bacteria has a lipid bilayer that undergoes rapid changes to adjust its fluidity to the changes in the environment.
- The outer membrane is asymmetrically composed of a special lipopolysaccharide providing less fluidity.
- Different serologic types of P. aeruginosa have been distinguished based on the evaluations of O-specific antigens.
- The organisms might appear pigmented as these can produce about six different types of pigments. Among these, the phenazine blue pigment is the most observed.
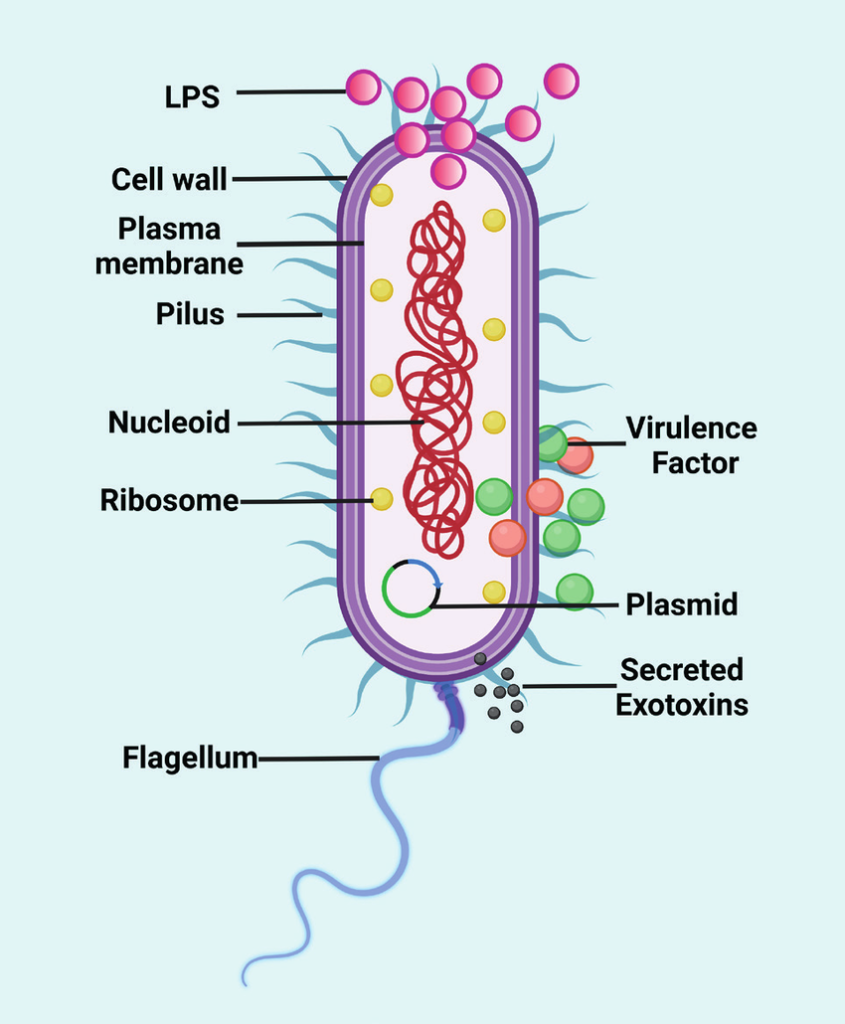
Fig 1: Diagram of Pseudomonas aeruginosa
Source: https://www.researchgate.net/figure/Schematic-diagram-showing-the-structure-of-the-Pseudomonas-aeruginosa-This-figure-has_fig2_371640522
Culture and Growth Characteristics
Usually, P. aeruginosa is aerobic in nature but can also grow anaerobically. Cultural medias used for the growth of P. aeruginosa is nutrient agar, 5% sheep blood or chocolate agar and MacConkey agar. Colonies observed are mucoid, pale and runny on MacConkey agar medium. It grows well in a medium containing carbon and nitrogen source and the optimum temperature in which it grows is 41°C. P. aeruginosa is resistant to most commonly used antibiotics and is resistant to high concentration of salts and heat. Cetrimide, acetamide and nitrofurantoin are added as a selective agent to culture medium to isolate P. aeruginosa from clinical and environmental samples.
Pseudomonas can be cultured on most general-purpose media and are identified with biochemical media and antimicrobial susceptibility testing. The growth on MacConkey agar shows pale, flat colonies with a spreading border. The colonies produced by P. aeruginosa are usually of three types;
- In clinical specimens, large, flat and smooth colonies with silver-grey metallic shining patches edges are observed.
- Similarly, in environmental samples the colonies observed are small, rough and convex.
- Mucoid type of colony is observed from respiratory and urinary tract infections.
- Produce green and fluorescing pigments.
Some of the colonies have a fruity odor and the occurrence of metallic patches.
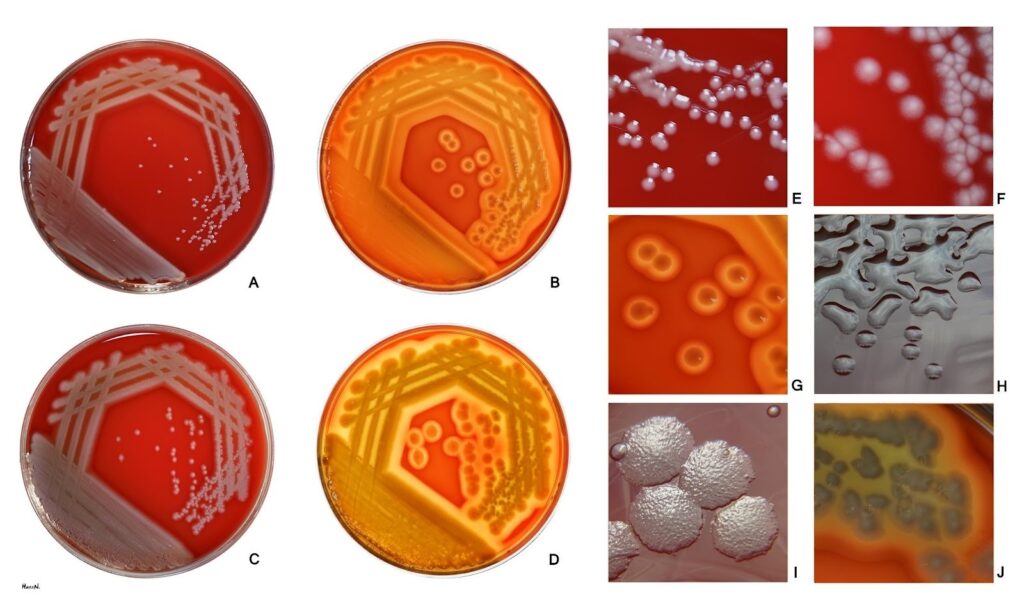
Fig 2: P. aeruginosa growing on Blood agar media
Image source: https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa#/media/File:Pseudomonas_aeruginosa_on_blood_agar.jpg
Pigments of P. aeruginosa
Pyocyanin, fluorescein, pyoverdin are a few pigments that are produced by P.aeruginosa which diffuses into surrounding media.
Pyocyanin:
- Bluish green phenazine pigment
- Soluble in chloroform and water
- Not produced by other species
- It is diagnostic of Pseudomonas aeruginosa
Pyoverdin (fluorescein):
- Greenish yellow pigment
- Insoluble in chloroform but soluble in water
- Produced by many other species
Pyorubin:
- Reddish brown pigment
- Insoluble in chloroform but soluble in water
Pyomelanin
- Brown to black pigment
- Production is uncommon
Biochemical and Identification tests
Table 2: Bio-chemical tests results
| Tests | Results |
| Motility | Positive |
| Hemolysis | Positive |
| H2S production | Negative |
| Indole | Negative |
| Oxidase Catalase | Positive Positive |
| Methyl redVPOxidative/FermentativeCitrateUreaseGlucoseMaltoseLactoseMannitolPyocyanin production Lysine decarboxylase Arginine dihydrolase | Negative Negative OxidativePositive NegativePositive Negative NegativePositiveVaries (+/-)Negative Positive |
Virulence Factors
Bacteria produce various virulence factors that help them to attach to host cells, damage tissues, evade the immune system and survive in the host body. Similarly, P. aeruginosa’s virulence factors are divided into three different types which are bacterial surface structures, secreted factors and bacterial cell-to-cell interaction. Each of them has a different role in the pathogenesis of P. aeruginosa. Some of the virulence factors and their roles in pathogenesis are described below.
Some of the virulence facter of P. aeruginosa are listed below:
- Lectin A (LecA/PA-IL): a sugar binding protein that binds galactose on host cells and helps bacteria to attach to epithelial cells. It disrupts epithelial barriers and is one of the important agents for biofilm formation.
- PlcB: known as Phospholipase C, a membrane destroying enzyme that breaks down the phospholipids in host cell membranes and helps in cell lysis, tissue invasion and promotes bacterial spread.
- Rhamnolipids: a biosurfactant glycolipids that is hemolytic in nature damages the host tissues, kills immune cells (neutrophils), disrupts cilia in respiratory tract and maintains motility.
- Exotoxin A (ToxA): it is like diphtheria toxin, inhibits protein synthesis in host cells and causes cell death and helps in severe tissue damage.
- Hydrogen cyanide (HCN): known as a toxic secondary metabolite that blocks cellular respiration. The role of this virulence factor is to inhibit cytochrome oxidase and it is mostly harmful in lung infections.
- Zinc metalloprotease known as LasB elastase targets elastin, collagen and fibrin and causes severe tissue destruction, also, inactivating immune system components.
Pathogenesis
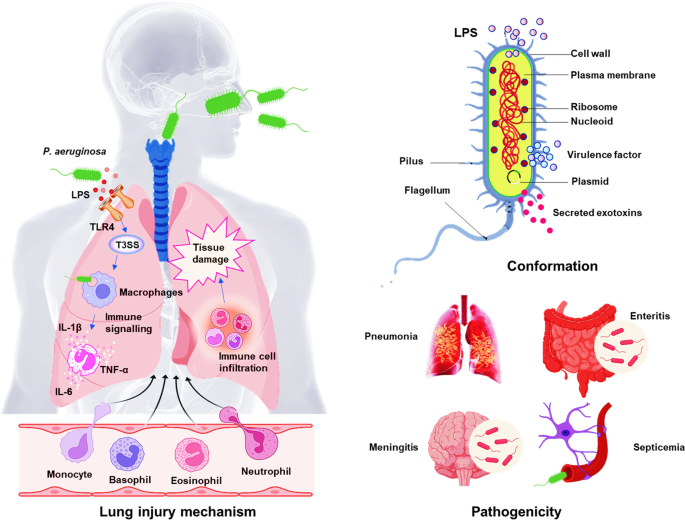
Fig 3: Diagram showing Pathogenesis of P. aeruginosa
Image source:
https://www.nature.com/articles/s41392-022-01056-1/figures/1
After the bacterial infection, when LPS activates TLR-4-dependent and independent inflammatory responses in the lung, epithelial cells secrete cytokines and chemokines which then recruits and activates both the innate and adaptive immune cells. Then some signs of inflammatory response activation are observed after the recruitment of neutrophils. Even if activation of neutrophils is critical for host defense, extremely activated immune cells infiltration causes severe tissue damage and worsen the bacterial infection.
Epidemiology and Transmission
It is a saprophytic bacteria that are found in natural sources. They are mostly found in moist environments of hospital wards, bathrooms, kitchen equipment, antiseptic and disinfectant solutions. Mainly, the P. aeruginosa have occurred amongst new-borns in nurseries and young infants in paediatric wards. Also, patients with immunosuppression, burned wounds and following cardiac and renal surgery are considered to be at high risk. The mode of infection occurs either from the environment or by cross-infection.
Clinical Manifestations
P. aeruginosa is one of the major causes of hospital-acquired infections. The common infections caused by P.aeruginosa are listed below:
- Respiratory tract infections: Immunocompromised patients and those who suffer from cystic fibrosis can be affected by P.aeruginosa, which causes sever hospital-acquired pneumonia. It can also cause chronic lung colonization.
- Ear infections: P. aeruginosa causes chronic otitis media (middle ear infection).
- Eye infections: The most common Pseudomonas infection in nonimmunocompromised patients is in the eye.
- Central Nervous system: In rare cases, P. aeruginosa can cause meningitis and brain abscess, mainly due to the hematogenous spread of the organism from infective pneumonia or UTI.
- Urinary tract infections: Most often, due to catheterization, instrumentation, and surgery, pseudomonal UTIs are acquired by the patients.
Laboratory Diagnosis
The laboratory diagnosis of P. aeruginosa are listed below:
Specimens: pus, wound swab, mid-stream urine, C.S.F., sputum and blood.
Microscopy: Gram Stain: Gram-negative rods (bacilli)
Process of Blood Culture (Aerobic):
- Blood (Adult: 5ml in 45ml, Paediatric: 0.5-1 ml BHI broth with 0.5% bile salt/ 8-10 ml for adults and 3-5 ml for children in Bactec Bottle is taken.
- Then, it is incubated aerobically at 35-37°C.
- After that, subculture on MacConkey Agar is incubated at 37°C for 24hrs.
- If NLF colonies which are pale in colour are observed then, oxidase and catalase are performed.
- When both oxidase and catalase are positive then we will further proceed in biochemical testing.
Bio-chemical tests: As for the biochemical tests, see table 2.
Antimicrobial susceptibility testing: According to the CLSI guidelines of 2025, commonly used antibiotics are Amikacin (AK), Gentamicin (GEN 10), Levofloxacin (LEV), Ciprofloxacin (CIP), Ofloxacin (OF), Piperacilin/ Tazobactem (PIT), Piperacilin (P), Ceftazidime (CAZ), Cefepime (CPM), Tigecycline (TGC), Polymixin B (PB), Collistin (C) and so on. Mullar-Hinton Agar is used for the AST.
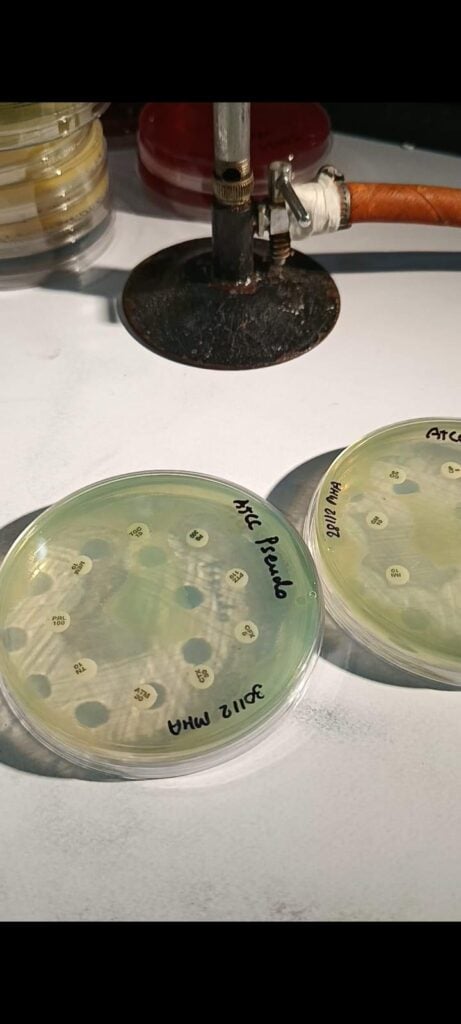
Fig 4: ATCC of Pseudomonas aeruginosa in MHA
Immunological and genetic Identification:
Various methods like Precipitation, Agglutination, Fluorescent antibodies, ELISA, RIA, Western blotting are used for immunological identification. As for the genetic identification, molecular techniques like Polymerase chain reaction (PCR), Amplified rDNA restriction analysis (ARDRA), PCR-SSCP (Single-Strand Conformation Polymorphism) and DNA fingerprinting are used. Here, we will be discussing the PCR techniques only. Commonly found genes in P. aeruginosa such as oprL, oprI, and toxA are targeted during PCR. The most commonly targeted gene is the 16S rRNA gene.
For its identification, following steps are followed:
- Pure culture is used for DNA extraction.
- Then, 16S rRNA is amplified by PCR.
- Gel electrophoresis is used to check the PCR product.
- Now, amplified DNA can be sequenced.
- BLAST is used for the comparison of known sequences in database and sequence.
- Presence of the expected band confirms P.aeruginosa.
The restriction pattern is shown by the P. aeruginosa helping to differentiate it from other Pseudomonas species.
Treatments
P. aeruginosa shows resistance to a variety of antibiotics, therefore susceptibility testing of isolates to antibiotics must be conducted in all cases.
Prevention and Control
- The major control measures should be used by healthcare professionals by strictly following hospital safety protocols.
- Regular monitoring and the disinfectants should be used in the wards where burned patients are kept.
Conclusion
In conclusion, P. aeruginosa holds the major clinical impact because of its virulence, persistence, and antibiotic resistance, mainly in vulnerable patients, therefore it is an important opportunistic bacterium.
References
Wilson MG, Pandey S. Pseudomonas aeruginosa. [Updated 2023 Aug 8]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK557831/
Qin, S., Xiao, W., Zhou, C. et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Sig Transduct Target Ther 7, 199 (2022). https://doi.org/10.1038/s41392-022-01056-1
Liao, C., Huang, X., Wang, Q., Yao, D., & Lu, W. (2022). Virulence Factors of Pseudomonas Aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Frontiers in cellular and infection microbiology, 12, 926758. https://doi.org/10.3389/fcimb.2022.926758
Iglewski BH. Pseudomonas. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 27. Available from: https://www.ncbi.nlm.nih.gov/books/NBK8326/
