Purines are heterocyclic aromatic compounds containing two fused rings. They are the most common nitrogen-containing heterocycles in nature. Purine is also present in significant amounts in red meat, beef, pig, dry peas, beans, lentils, and oats.
The purine ring is assembled from a variety of precursors i.e., glutamine, glycine, aspartate, and N10– formyl tetrahydrofolate.
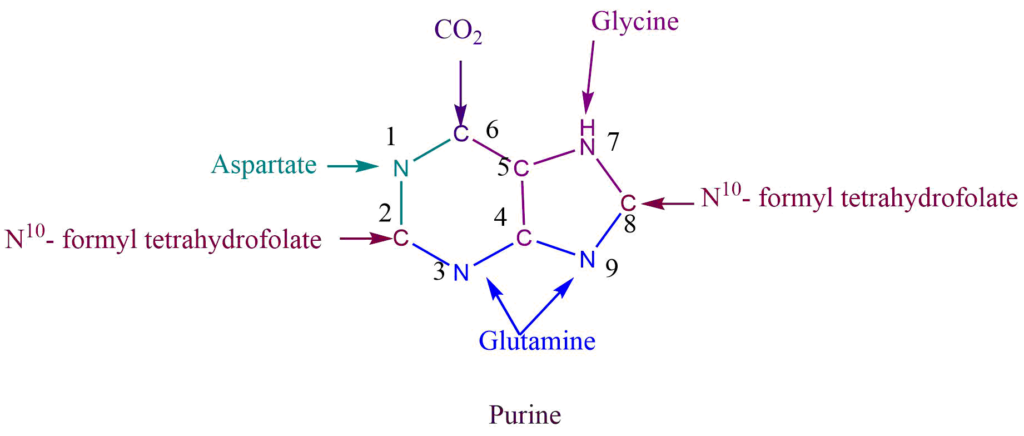
N1 of purine come from the aspartate while the amide group of glutamine provides N3 and N9. C6 of purine comes directly from CO2, C2, and C8 from N10– formyl tetrahydrofolate, while C4 and C5 come from glycine.
Interesting Science Videos
De-nevo synthesis of purine nucleotide
In the de novo pathway Using 5-phosphoribosyl-1-pyrophosphate (PRPP), enzymes build purine and pyrimidine nucleotides using simple molecules such as CO2, amino acids, and tetrahydrofolate. De-nevo synthesis of purine nucleotide begins with simple starting materials such as amino acids and bicarbonate. Unlike the case for pyrimidines, the purine bases are assembled already attached to the ribose ring.
Reaction Steps
Ribose 5- phosphate of carbohydrate metabolism is the starting material for purine nucleotide synthesis. Ribose 5- phosphate reacts with ATP to give phosphoribosyl pyrophosphate.
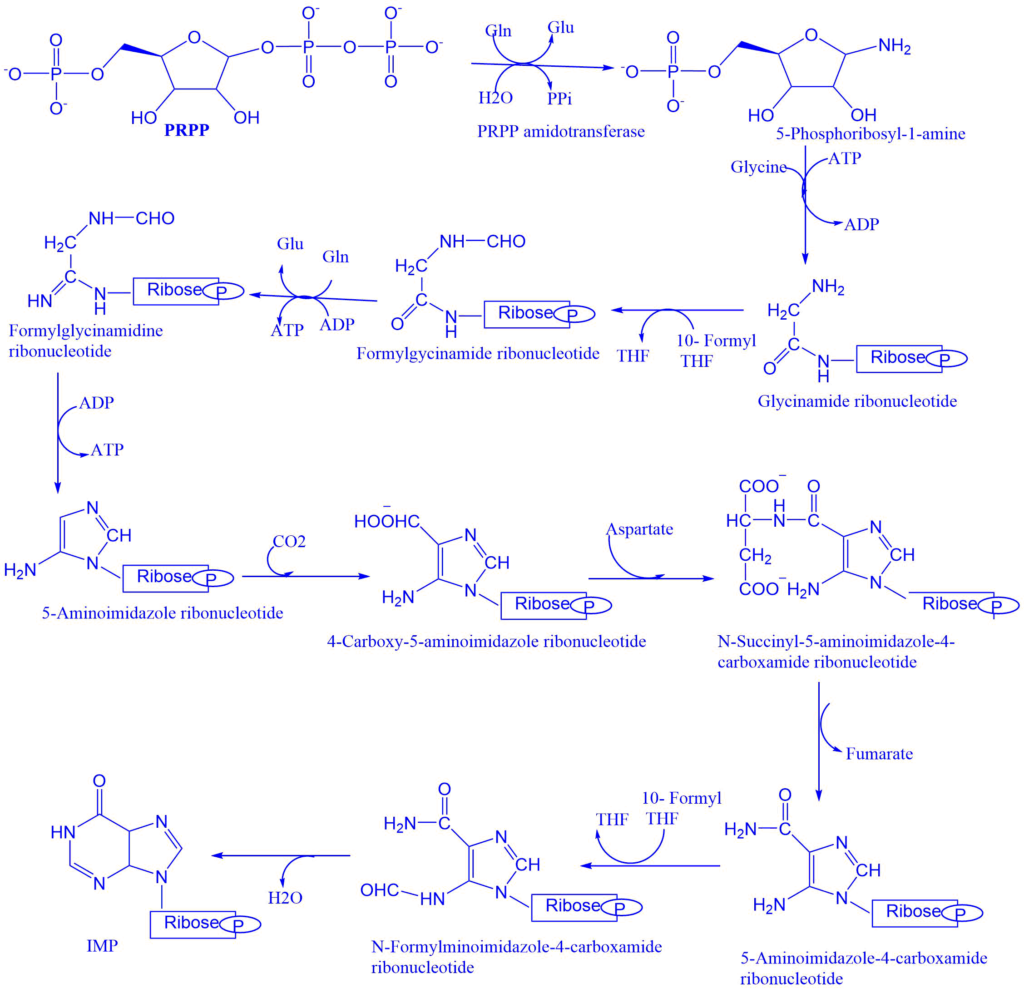
- Glutamine transfer amide nitrogen to PRPP which replaces the pyrophosphate and produces 5-phosphoribosyl-1-amine. The enzyme PRPP glutamyl amidotransferase catalyzes this reaction step. The feedback inhibition of IMP, AMP, and GMP controls this reaction. This is committed reaction.
- The carboxylate group of activated glycine is coupled to the amino group of phosphoribosylamine to form a new amide bond. That leaves the amino group of glycine free to act as a nucleophile in the next step.
- After activating the formate, it is added to the amino group to form formylglycinamide ribonucleotide. The formyl group is transferred from N10-formyltetrahydrofolate by one enzyme, while the formyl phosphate formed by the other enzyme is directly added to the glycine amino group.
- Inner amide group is activated and then converted into amidine by the addition of ammonia derived from glutamine.
- Formylglycinamidine ribonucleotide cyclizes to form the five-membered imidazole ring found in purines.
- The exocyclic amino group attacks the activated bicarbonate. The product of the reaction rearranges to transfer the carboxylate group to the imidazole ring.
- Aspartate condenses with the aminoimidazole carboxylate ribosyl 5- phosphate to form 4-succinylcarboxamide ribosyl 5-phosphate. The enzyme synthetase catalyzes this reaction step.
- 4-succinylcarboxamide ribosyl 5-phosphate eliminates fumarate, from 5 aminoimidazole-4-carboxamide ribonucleotides. The formyl group from N10 formyltetrahydrofolate adds to the nitrogen atom to form N-formylaminoimidazole-4-carboxamide ribonucleotide. Then the enzyme cyclohydrolase catalyzes the ring closure reaction with the elimination of water, to give the final product i.e., inosine monophosphate (IMP). Inosine monophosphate (IMP) serves as the parent purine nucleotide for the synthesis of other purine nucleotides. Adenosine monophosphate (AMP) and guanosine monophosphate (GMP) were formed from IMP.
Formation of AMP and GMP from IMP
Conversion of IMP to either AMP or GMP utilizes two step energy-requiring pathway. The synthesis of AMP requires GTP as an energy source whereas GMP requires ATP.
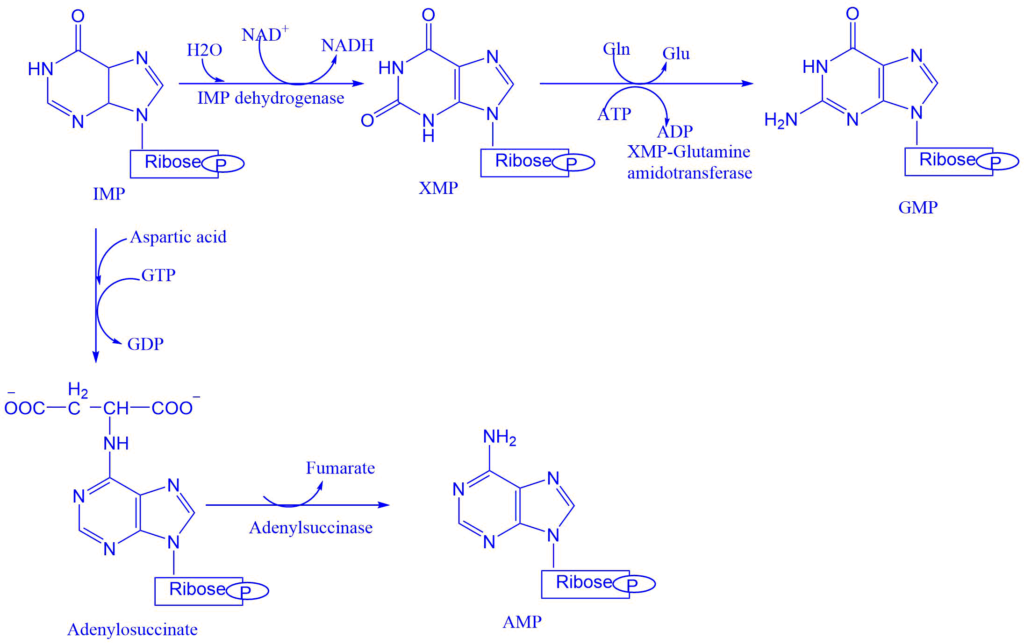
- Adenylsuccinate synthase catalyzes the conversion of IMP into adenylosuccinate. This eliminates the fumarate to give the final product i.e., adenosine monophosphate (AMP).
- IMP dehydrogenase catalyzes the formation of xanthosine monophosphate from IMP. Which is then converted into guanosine monophosphate (GMP). The enzyme XMP-glutamine amidotransferase catalyzes this reaction step.
Conversion of nucleoside monophosphate to the nucleoside diphosphate and triphosphate
Nucleoside monophosphate kinase converts the nucleoside monophosphate into nucleoside diphosphate (NDP). ATP is generally the source of transferred phosphate because it is present in higher concentrations.
E.g., Adenylate kinase: AMP+ ATP ⇌ 2ADP
Gyanolate kinase: GMP+ ATP ⇌ GDP + ADP
Nucleoside diphosphate and triphosphate are interconverted by nucleoside diphosphate kinase.
GDP+ ATP ⇌ GTP + ADP
CDP+ ATP ⇌ CTP + ADP
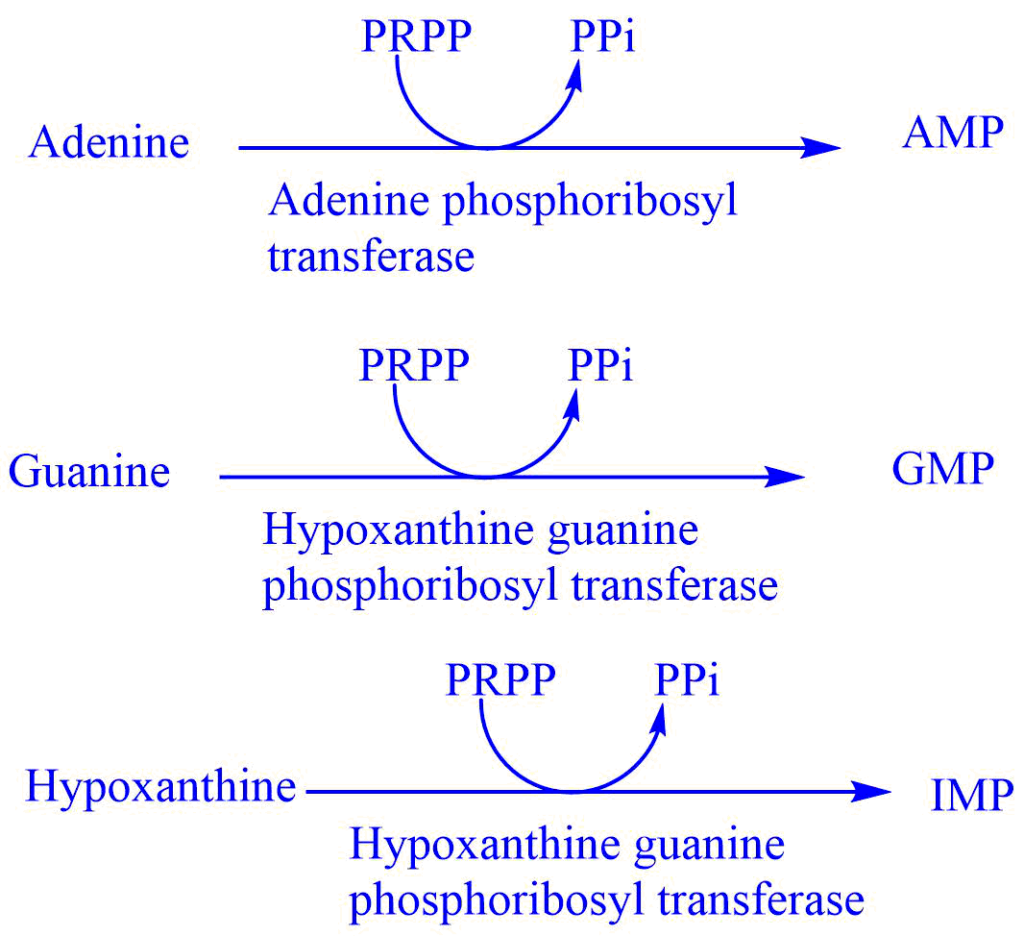
Salvage pathway of purine synthesis
Purines that result from the normal turnover of nucleic acids or that are obtained from a diet not degraded can be reconverted to nucleoside triphosphates and used by the body. This is referred to as the salvage pathway of purine.
This pathway involves two enzymes adenine phosphoribosyl transferase (APRTase) and hypoxanthine phosphoribosyl transferase (HGPRT). HGPRT is the starting material in the salvage pathway and acts as the source of ribose 5-phosphate. The salvage pathway is important in some tissues where the de-nevo synthesis of purine nucleotide is not operative.
Regulation of purine synthesis
1. The intracellular concentration of PRPP regulates purine synthesis. This in turn depends on the availability of ribose5-phosphate and PRPP synthetase.
2. Purine nucleotides control the PRPP glutamyl amidotransferase by the feedback mechanism. That is if AMP and GMP are available in an adequate amount to meet the cellular requirement, their synthesis is turned off at the amidotransferase reaction.
3. The other step of regulation is in the conversion of IMP to AMP and GMP. AMP inhibits adenylosuccinate synthetase whereas, GMP inhibits IMP dehydrogenase. So, AMP and GMP control their synthesis from IMP by the feedback mechanism.
Importance of purine nucleotides
1. They are the building blocks of nucleic acids (like DNA and RNA)
2. They are components of coenzymes (FAD, NAD, and Coenzyme A)
3. cAMP and cGMP participate in the regulation of many metabolic pathways.
4. ATP and GTP act as the source of biochemical energy.
Degradation of purine nucleotides
Purine nucleotides are sequentially degraded by the removal of portions of the nucleotides. The end product of purine catabolism in humans is uric acid.
The sequences of reactions involved in the degradation of purine degradations are:
1. The nucleotide monophosphates (AMP, IMO, and GMP) are converted to their respective nucleoside form (Adenosine, Inosine, and guanosine) by the reaction of nucleotidase.
2. Removol of amino group either from AMP or adenosine produces IMP or inosine respectively.
3. Inosine and guanosine converted to their respective purine bases, hypoxanthine and glutamine.
4. Glutamine undergoes deamination by guanase to form xanthine whereas hypoxanthine undergoes oxidation by xanthine oxidase to form xanthine.
5. Xanthine oxidase further oxidizes xanthine into uric acid.
Uric acid is the final excretory product of purine metabolism in human beings. Uric acid can serve as an important antioxidant by getting itself converted to allantoin. Animals other than primates oxidizes uric acid by an enzyme uricase to allantoin. Which is then converted into allantoic acid and excreted in some fishes. Allantoic acid on further degradation produces urea (in amphibians, most fishes, and molluscs), and later on ammonia (IN marine invertebrates).
Disorders of purine metabolism
Gout
Gout is characterized by elevated uric acid concentration in blood (hyperuricemia) and urine due to various metabolic abnormalities which lead to the overproduction of purine nucleotides through a de-nevo pathway. In severe hyperuricemia, sodium urate crystals deposit in joints. This causes inflammation, which leads to gouty arthritis.
Pseudo gout
The clinical symptoms of pseudogout are similar to those of gout. However, this is results due to the deposition of calcium pyrophosphate crystals in joints. Furthermore, serum uric acid concentration is normal in pseudogout.
Lesch-Nyhan syndrome
This is genetic disorder, results due to the deficiency of salvage enzyme hypoxanthine guanine phosphoribosyl transferase. Children with this deficiency display compulsive self-destructive behavior. They also show the sigh of mental retardation.
Suggested video
References
- Smith, C. M., Marks, A. D., Lieberman, M. A., Marks, D. B., & Marks, D. B. (2005). Marks’ basic medical biochemistry: A clinical approach. Philadelphia: Lippincott Williams & Wilkins.
- Satyanarayana U. & Chakrapani U. (2013). Biochemistry (4th ed.). Elsevier Health Sciences APAC.
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3243375/
- https://www.pharmaguideline.com/2007/01/biosynthesis-of-purine-and-pyrimidine.html
- https://www.slideshare.net/YESANNA/biosynthesis-of-purine-nucleotides
- https://bioone.org/journals/the-arabidopsis-book/volume-2002/issue-1/tab.0018/Purine-and-Pyrimidine-Nucleotide-Synthesis-and-Metabolism/10.1199/tab.0018.full
- https://slideplayer.com/slide/13110257/
- https://library.med.utah.edu/NetBiochem/pupyr/pp.htm
