Toxicity analysis is crucial in the drug discovery and development process to assess the safety of a drug candidate and identify the adverse effects of the drug candidate, such as genotoxicity, immunotoxicity, carcinogenicity, and so on.
Our exposure to chemicals is increasing as the number of chemicals and their combinations as mixes grows exponentially. Our daily lives involve constant interaction with chemicals as we live in a highly reactive chemical environment that is present in everything from the food we eat to the medications we are prescribed to the cosmetics we wear to the air we breathe. However, the amount and duration of chemical exposure can be both detrimental and beneficial. As a result, the hazardous potential of the compounds and their combinations must be experimentally validated.
Interesting Science Videos
Importance of In-Silico Toxicity Analysis
Due to several constraints such as time, expense, and ethical considerations with animal studies, it is impractical to test all chemical compounds on experimental platforms. Thus, in silico toxicity analysis is rapidly growing as a key platform for predicting the toxicity of compounds that may be detrimental to humans, animals, plants, and the environment. To predict the toxicity effects of chemicals and reduce the time, expense, and requirement for animal testing, in silico toxicity models are aimed at enhancing current in vitro toxicity approaches.
Using the in silico approach, a toxicity analysis of compounds can be quantified in terms of toxicity endpoints such as mutagenicity, carcinogenicity, and many more. It can also be quantified numerically, such as LD50 (lethal dosage) values, and qualitatively, such as binary (active or inert) for certain cell types, tests, or indication areas, such as cytotoxicity, immunotoxicity, and hepatotoxicity.
Software/ Web Servers used for Toxicity analysis
LIVERTOX
LIVERTOX is an important website for toxicity analysis that delivers up-to-date, accurate, and easily accessible information on the diagnosis, etiology, frequency, patterns, and treatment of liver impairment caused by prescription and nonprescription medicines, herbs, and dietary supplements. The website is primarily intended for physicians and healthcare workers who may see patients with drug-induced liver impairment on a rare basis, such as family practitioners, internists, pediatricians, psychiatrists, surgeons, specialists, and subspecialists in all fields of medicine. LIVERTOX also incorporates a case registry, which will allow for more scientific study and definition of the clinical patterns of liver injury.
TOPKAT: Toxicity Prediction by K(C)omputer Assisted Technology
TOPKAT is a computer-based method for predicting chemical carcinogens was assessed based on its ability to predict the carcinogenicity of compounds.
TOPKAT analyses the toxicity of compounds based on their two-dimensional (2D) molecular structure. It utilizes cross-validated QSTR models to evaluate specific toxicological outcomes. Verified databases, information-rich descriptors, highly predictive QSAR-based models, and prediction validation methodologies enable users to examine the model’s relevance to the molecules under assessment.
TOPKAT employs descriptors to measure chemical features such as molecule bulk, shape, and symmetry, as well as information-rich electro-topological descriptors (E-state). This E-state descriptor quantifies the electrical and topological properties, which are subsequently utilized to characterize interaction on the site.
LAZAR: Lazy Structure-Activity Relationships
The LAZAR (ARI) system is a free online tool for toxicoinformatics. This method is used for toxicity analysis of chemical compounds accurately by using an inductive database with experimentally obtained toxicity data. LAZAR’s prediction capacity relies heavily on high-quality data input into its inductive database.
HazardExpert
HazardExpert is an (ARI) expert system. The software’s open architecture allows chemists, toxicologists, drug disposition experts, and environmental managers to change and optimize the data used for toxicity estimation. Chemical structures can be retrieved from a database or manually entered by the user for prediction purposes. The user is required to specify the species, dose amount, method, and duration of exposure.
OncoLogic™
The OncoLogic™ model is an expert system that replicates human judgment by following knowledge rules based on studies of how chemicals cause cancer in both animals and people. OncoLogic™ uses user-provided chemical and use information to estimate the chemical’s carcinogenic risk based on established knowledge principles.
TEST
The Toxicity Estimation Software Tool (TEST) was created to help users assess the toxicity of compounds using Quantitative Structure Activity Relationships (QSAR) methods. QSARs are mathematical models that predict toxicity levels based on physical features of chemical structures (also known as molecular descriptors). The TEST software estimates the toxicity and physical attributes of substances based on their molecular structure.
TEST allows users to assess toxicity without the need for additional programs. Users enter a chemical to be evaluated by sketching it in the included chemical sketcher window, typing it into a structure text file, or importing it from the included structure database.
AMBIT
AMBIT is an open chemoinformatic system meant to assist companies by facilitating chemical safety assessments. AMBIT also provides read-across and category generation of chemicals, which are critical approaches in chemical safety assessments.
The AMBIT web services package was created as an expansion of AMBIT modules, adding the ability to generate (Quantitative) Structure-Activity Relationship (QSAR) models and an OpenTox API-compliant interface. The way data and processing resources are represented in the W3C Resource Description Framework makes it easier to integrate them as Linked Data. Uploading datasets with chemical structures and an arbitrary set of characteristics makes them instantly available online in a variety of forms.
Tox Tree
ToxTree is an in-silico, non-testing method for assessing the risks and hazards of a chemical (http://toxtree.sourceforge.net/). It assigns a Cramer class (I, II, or III, from possibly least to most harmful) by using a decision tree to apply physicochemical and structural principles to information about the compound’s properties and structure. Each class has a toxicological concern threshold (TTC), which is calculated using human exposure in micrograms per day. Comparing the compound’s TTC to actual/predicted exposure levels tells whether the risk level is acceptable. ToxTree also predicts skin and eye irritation, mutagenicity/carcinogenicity, biodegradation, skin sensitization, and protein/DNA binding warnings. It is largely concerned with predicting toxicity-related effects and investigating the toxicological profile of chemicals.
ProTOX
ProTox is a web server for predicting the toxicity of small compounds. It accepts numerous chemical input formats, including generic names, structures, and chemical formulas, and then classifies the chemical using the program’s reference database. This model evaluates the predicting “oral” toxicity in rodents and will produce several possible outcomes.
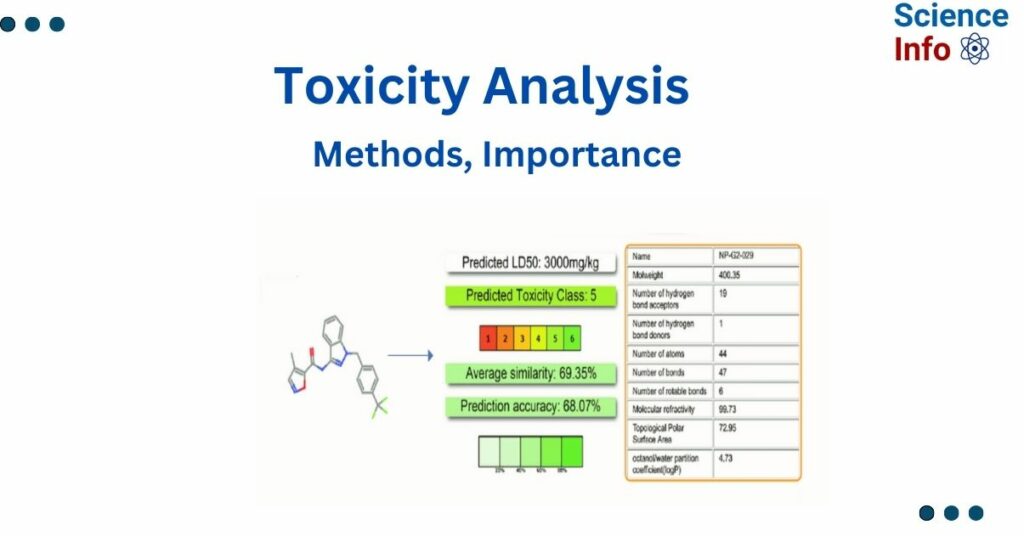
The findings on this will display the projected median lethal dose (LD50) in mg/kg weight, toxicity class, prediction accuracy, average similarity, and the three most similar dangerous chemicals from the dataset with known rodent oral toxicity values. If projected toxicity target information is available, it will be displayed along with the target’s name, average fit, and similarity of the input molecule to the pharmacophore and known ligands of the relevant targets.
ProTox-II is an in-silico approach that determines the toxicity class of different compounds and classifies them as class 1 and 2 (fatal), class 3 (toxic), class 4 and 5 (harmful), and class 6 (non-toxic) based on LD50 value. The ProTox website contains both chemical and molecular target knowledge. The ProTox-II webserver is unique in that the prediction scheme is divided into different levels of toxicity, such as oral toxicity, organ toxicity (hepatotoxicity), toxicological endpoints (such as mutagenicity, cardiotoxicity, cytotoxicity, and immunotoxicity), toxicological pathways (AOPs), and toxicity targets, providing insights into the possible molecular mechanisms underlying such toxic responses.
References
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6031011/
- https://www.ingentaconnect.com/content/ben/mrmc/2007/00000007/00000005/ art00006.
- https://academic.oup.com/nar/article/46/W1/W257/4990033
- https://www.nature.com/articles/s41598-022-18327-0
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3120779/
